Quick Chat: Professor Dr. Brian Ingalls
EMMA LINDSAY - AUGUST 10, 2022
Dr. Brian Ingalls is a professor at the University of Waterloo in the Department of Applied Mathematics. He is also cross appointed to the Departments of Biology and Chemical Engineering. Dr. Ingalls’ research is focused on synthetic and systems biology. His lab is the Ingalls Quantitative Cell Biology Lab at the University of Waterloo, where his group utilizes mathematical and computational tools to build and analyze kinetic models of biomolecular systems. Their focus is on intracellular networks and heterogeneous microbial communities. Currently, their projects are centered on model-based design of synthetic bacterial gene regulatory systems.
How did you get into synbio?
When I was finishing my PhD, I was working with feedback control systems and my advisor pointed me to some recent stuff that was being done in reverse engineering a biochemical regulation. I went to a talk by Michael Elowitz, who presented the repressilator. I had no biology up to that point, and I was just blown away that it was possible to do that kind of engineering project in that space. And then I got lucky, and I ended up in a postdoc group doing engineering and biology.
What are some of the projects currently going on in your lab?
Bacteria mediated cancer therapy. This is something that most people have never heard of. In the mid 1800’s, it was observed that sometimes cancer patients got bacterial infections and if they survived, sometimes it helped with their cancer. People played around with it, but they did not really understand what was happening. And then, chemotherapy and radiotherapy were developed, and it fell off the radar because people had better ways to address cancer. It's come back a little bit, people are renewing their interest in it because of the means we have now to manipulate the bacteria doing this. The basic idea is that anaerobic bacteria solve in a particular way the targeting problem. In cancer therapy, one of the biggest problems is targeting cancer cells, and not targeting healthy cells. The feature that bacteria mediated cancer therapy was taking advantage of is that a lot of cancers produce these solid tumors, and these solid tumors have trouble being properly oxygenated, so you end up with this mass of cells that have poor access to oxygen. This produces an anaerobic environment inside the person's body. So, if you have cells or spores of anaerobic bacteria, you introduce them into a person. Those cells, if they find their way into the anaerobic core of that tumor, can proliferate and propagate. And if they don’t, they just get cleared since they are inert. So that is an interesting way of solving this targeting problem. People have been looking at this over the last couple of decades in terms of engineering those cells so that they can do different things. Naturally, as they grow, they can chew out some of the cancer cells in the area and elicit an immune response because there's this infection going on so that can help with the cancer remission. But also, you can engineer the production of drugs or prodrugs. We’re looking at the Clostridium species and we're doing some engineering with that to do two things: one is to introduce a quorum sensing mechanism so that the activity can be turned on when they reach a significant population density in this tumor environment and have targeted activity in particular areas; the second thing is we are also looking at increasing their air tolerance so that they can proliferate a little more than they normally could in that environment to have the effect that we need.
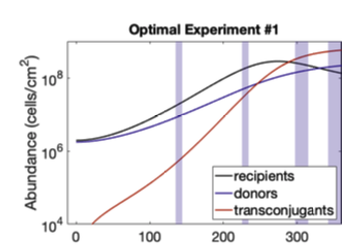
Optimal experimental design. It was established one hundred years ago in the early 20th century for linear descriptions on how experiments depend on the set up of the experiment. The basic idea is that if you have a particular objective for your experiment and you have a limited set of resources that you can expand your project on, then which resources should you choose to address the goal of understanding the system in a particular way? In the linear space this is a nice theory and applies when you make these assumptions about a system's behaviour linearity, which is not typically the kind of thing you see in biomolecular networks. If you take the same theory and try to apply it to more complicated linear scenarios, for example dynamic systems, then you have the same kind of questions. Which experiments should I do? One basic question first is if you’re taking a time series, where do you choose your time points? That's the kind of questions that one can address. So recently we’ve developed tools and some software packages along those lines that essentially allows folks to do that kind of analysis.
An example of the optimal experimental data. Graph by Nathan Braniff, PhD Candidate
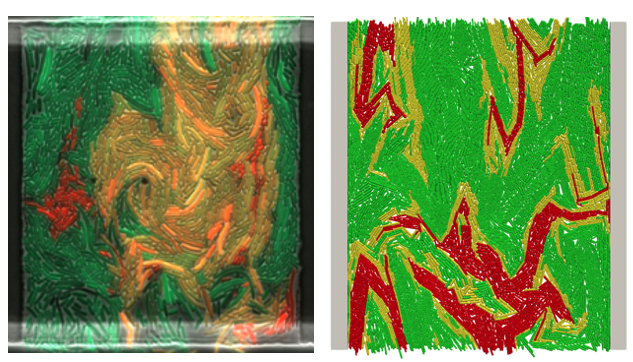
Behaviour of bacterial communities. We're looking at baby versions of bacterial ecology. Bacterial ecology is very complicated and it’s very difficult to characterize what is going on in those systems. What we do in the lab is we set up very simple communities and start with characterizing their behaviours and working towards things that are more complicated. We are looking at things like horizontal gene transfer, phage infections, and more generically inhibition in terms of a diffusible toxin release from cells or a contact dependent effect. The tool we have for characterizing these systems is time lapse fluorescence microscopy. We have a set up that allows us to image the entire population in a single focal plane and we add fluorescent markers to these cells to indicate their state. We can run these experiments for twenty-four hours or more and we can watch the cells propagate and interact. What we have alongside that data is the characterization of the data in terms of these dynamic models. One opportunity is to use this modeling structure to understand how these systems behave, for instance to infer things like conjugation rate, growth rate, and effective conjugational growth. The goal here is to eventually use these models for model-based design of manipulation of these systems. These systems are in lots of different environments and being able to manipulate them sounds promising.
Image by Aaron Yip, PhD student.
You use mathematical and computational tools for your projects. Are these techniques common in synthetic biology?
The main mathematical tool the group uses is kinetic modeling to characterize system behavior and explore possibilities. That can be either the reverse engineering of systems to better understand what is there, or from a forward engineering perspective, model-based design. We can build models with some degree of accuracy in terms of prediction and that’ll allow us to explore design space much more cheaply than you could running batches and batches of experiments. I would say it's fairly common. There are lots of people that go into life science and have an aversion to computation. That's changing. There was a time when a lot of people in biology spent their whole careers and did very little computational work. The human genome project was a breakthrough for bioinformatics being identified as a key part of molecular biology. This kind of work is a bit more esoteric. Standard bioinformation is about things like genome analysis, sequence alignment, working with static data, and there's a lot of that data available. This kind of work requires more data to put these kinds of descriptions together. That kind of data is hard to collect, it is expensive to run these experiments so it's often not the first thing people do. I like to think it is becoming increasingly more common, but I wouldn't say it's very common. But there are lots of projects where it's an important piece and it's becoming a more valuable tool as people recognize that the data is available. The basic idea is that the systems are very complicated so in every other branch of science when you hit a really complicated system you need to turn to these tools to understand.
Are there any resources that Canada needs to help strengthen the field of synthetic biology? Where do you think Canada can improve?
Canada's economy is based on resource extraction which comparatively doesn't require a lot of R&D. That's been the basis for the economy since the country was founded. That is in part what has led to a sort of risk aversion in terms of investment, both private and public, so Canada is often lagging when it comes to new technologies and risky technologies. We do have certain strengths for sure. The educational system is great at producing people who can do this sort of work, and who are interested in this sort of thing. I think there's a lot of promise, and there certainly could be more delivery on that promise if we had more investment.
Are there any applications or techniques in the synthetic biology field that you’re excited for?
There's a lot of short term promise in biomanufacturing. Improving biomanufacturing is an iterative improvement, and so that does seem to be an area that the community is continuing to embrace as where there will be lots of impacts of synbio. One of the basic questions is about the release of organisms. So, if you're going to make an organism and you're going to use it, you have to set it free into the environment. People are understandably nervous about that. There are lots of discussions, and I don't think there's going to be any problem there. If you can have your advantage in the lab or the factory, and you never have to have that discussion, then it makes life a lot easier.
Do you have any advice for someone that wants to get involved in synthetic biology?
I could say two things. One is networking. There are groups and communities such as iGEM, and there’s always an opportunity to reach out. There are lots of opportunities to get involved even at the high school level. That’s something people should keep in mind. And the community is very welcoming and happy to have people interested at all stages. The other thing I would suggest in planning an education that could lead in this direction is being open and trying to embrace ideas across disciplines. A lot of educational programs are very siloed and disciplinary. Especially as an undergrad, that's how the program is set up and that's what you do. You take the courses you need to take. Seek out opportunities to expand that toolbox. Given that synbio is a relatively young field and a very interdisciplinary field, the best positioning one can have as a junior researcher, or someone interested in entrepreneurship is to get exposure to as many as those things as possible. So do a little bit of computing or entrepreneurship or life science or bioinformatics. Often one must be proactive about reaching out cause standard programs are typically very siloed and disciplinary.
Dr. Ingalls’ research in developing mathematical models is important for the field of synthetic biology. These models can help predict behaviours of intracellular networks and cellular communities, help build an understanding of complicated systems, and help further the progress of synthetic biology research